Research Interests
Recent years have witnessed the success of applying sequencing technologies to accelerate crop biological studies and breeding. Today, there is an urgent need for crop improvement as global population growth demands a substantial increase in crop productivity in the next few decades, while consumers tend to have higher expectations on food quality. Stemming from this, our research focuses on:
Using comparative genomics approach to understand genetic basis of crop domestication.
Achieving crop breeding success on both yield and quality lies in the knowledge and availability of key causative alleles. Fortunately, identification of such alleles can be greatly facilitated by genomic and population genomic studies. Our previous and current work has beening digging deep into the genome biology of different systems, and important discoveries from that have provided a foundation for our future research, in which we will use comparative genomics approach to address two fundamental questions: 1) how genes and their regulatory elements work together to affect phenotypic outcomes, and 2) how such mechanisms could be deployed for crop improvement.
Seeing through the dark matter in the plant genomes
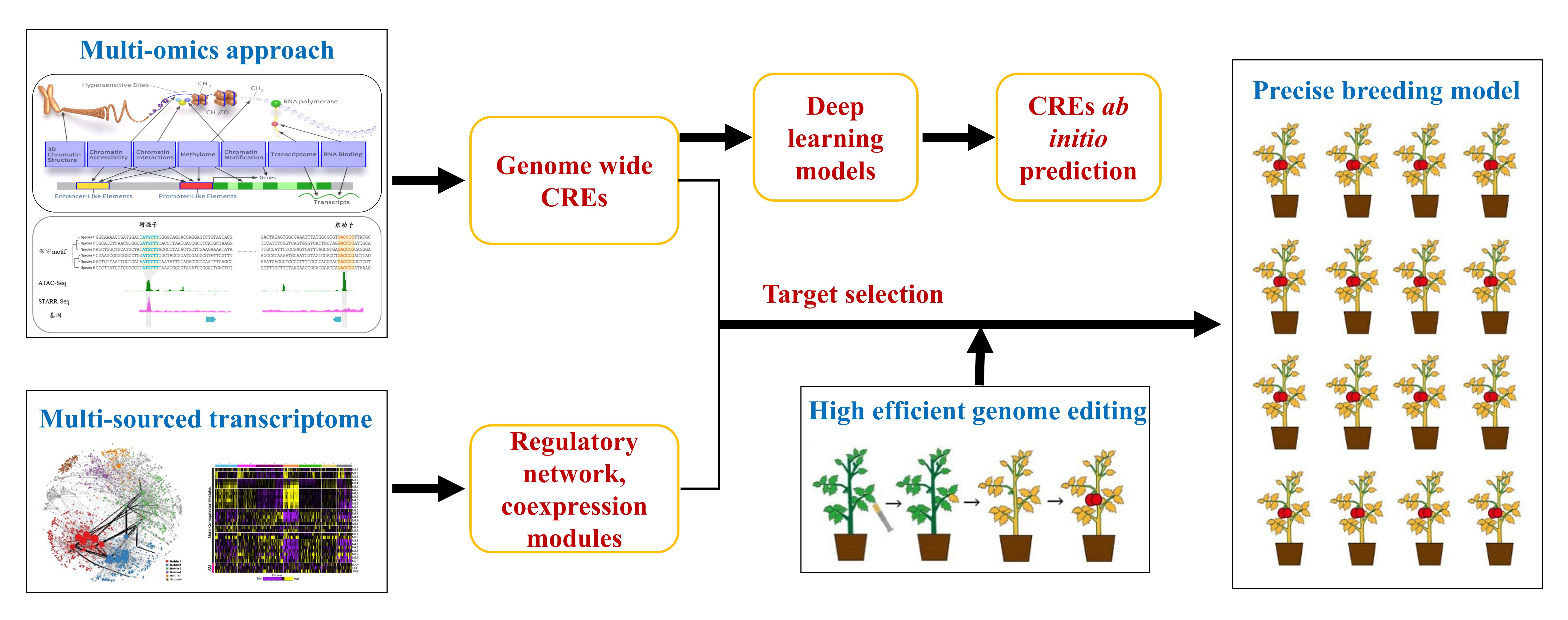
Plant genomes consist of an overwhelming part of non-coding regions where cis-regulatory elements (CREs) reside in. Identification and functional characterization of CREs in the dark recesses of plant genome is a daunting but essential task, in that: 1) most of causative variations responsible for the agronomic traits are located in non-coding regions, in which CREs are the hotspots for genetic variation; and 2) CREs and upstream ORFs are often considered as the ideal targets for genome editing, especially when creating a continuum of phenotypes is favored. A complete picture of CREs in plants is instrumental to the understanding of the transcriptional regulation in individual species and the plasticity of gene regulatory networks on an evolutionary scale. Research on plant CREs is still in its infancy. The short-term goal of my lab on this project is to understand the pattern and diversity of CREs in green plant lineage with a particular focus on characterizing functional CREs in the selected major crops, whereby the long-term goal is to scale up taxonomical coverage and develop a unified framework for comparative analysis of plant CREs. Expected results on the identification and analysis of plant CREs would greatly facilitate crop breeding.
Genome editing in flowering trees
Genome editing in flowering trees is often less efficient than in annual plants. This project aims to develop new strategy to accelerated genome editing in horticultural important tree plants.